Leukocyte counts in capillary blood micro-samples based on targeted analysis of DNA methylation with digital droplet PCR - “Prick-Me”
In clinical routine, the analysis of the leukocytes composition in peripheral blood represents a daily procedure. Cell counting is usually performed manually or automatically, however, all conventional methods require fresh blood samples (not older than 24 hours). Alternatively, it is possible to estimate the cellular composition based on epigenetic modifications, which are acquired during hematopoietic differentiation.
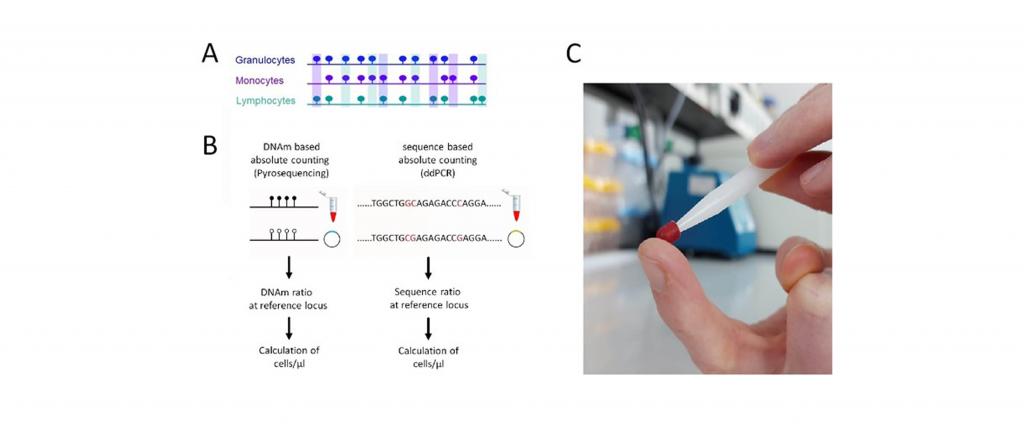
In our previous work, we demonstrated the possibility to measure DNA methylation (DNAm) at cell-type-specific CG dinucleotides (CpGs) via “pyrosequencing” to estimate the absolute and relative composition of white blood cells (WBC). Epigenetic cell counts are applicable to frozen blood samples and small volumes of blood (about 30 µl).
The “Prick-Me” proposal aims to further develop and validate this application for capillary blood, which can be collected by a simple finger prick. For quantitative analysis, the so-called “droplet digital PCR (ddPCR)” shall be used, providing more reliable results than pyrosequencing. This procedure will facilitate regular measurements and self-collection at home by the patients themselves.
Project partner: VIP+ Bundesministerium für Bildung und Forschung (Validierungsförderung VIP+)